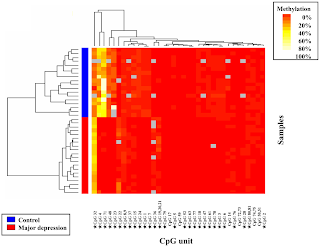
In the August 30th issue of PLoS One, Fuchikami and colleagues report their findings about a new biomarker of severe depression based on Brain-Derived Neurotrophic Factor (BDNF) gene methylation profiles (reference). Briefly, the authors analyzed the methylation profile of the BDNF gene in blood samples collected from 20 clinically diagnosed severe depression patients and 18 healthy human volunteers (see figure 1). Their analysis covered 81 CpG units upstream of exon 1 (CpG I) and 28 CpG units upstream of exon 4 (CpG IV) of the BDNF gene. Differential methylation status in CpG I appeared markedly different between patients and controls, with an overall trend for hypo-methylation in patients with major depression. The biological implication of this methylation profile is currently unknown.
Fig.1
Considering the small sample size used in this study, these findings should be viewed as an initial screening for potential biomarker candidates which will require substantially more work to be confirmed. First, because the individuals enrolled in this first study were exclusively of Japanese origin, the relevance of BDNG gene methylation status as a biomarker of depression remains to be established in a more ethnically diverse population. Second, as I have mentioned in an earlier post (link), the reductionist sample selection process used in this study probably yielded over-optimistic statistical association values that may not translate well to the more complex real-world. Indeed, the diagnosis of major depression is almost never made as a simple binary determination of “healthy” vs. “depressed”. Rather, the diagnosis of depression is a process of eliminating other conditions that manifest themselves with similar symptoms. Hence, analysis of the BDNF gene methylation profile in clinically related psychiatric conditions should constitute an important follow up to this initial study. Finally, assuming that these biomarker candidates are confirmed, it would be particularly interesting to determine whether current treatments for depression affect the methylation profile of the BDNF gene.
Of note, it seems that the field of biomarker discovery in the area of depression is picking up speed lately. This paper comes one day after the announcement by Lundbeck Canada of a $2.7 million donation in support of biomarker discovery in the area of major depression and bipolar disorder (announcement), and a few weeks after the cover story of Ridge Diagnostics’ depression blood test in the August issue of Psychiatric Time (see earlier post).
Thierry
Sornasse for Integrated Biomarker Strategy